IBD Estimation
The objective is to compute the probability of two alleles being shared IBD given the genotype data at that locus and the entire, unbroken pedigree. With very deep pedigrees it is often the case that many generations will have no genotype data, whereas more recent generations do. An efficient implementation will necessarily take this difference in information into account and execute different strategies in the two realms. My approach is based on the same recursive method suggested by Wang et al. (1995) and Davis et al. (1996) and is analogous to the recurrence relation for kinship coefficients,
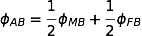
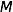
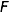
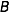
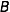
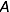
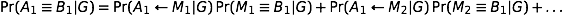
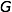

Accounting for missing data correctly is critical to any real-world data analysis. Both Wang et al. and Davis et al. suggest Monte Carlo approaches for dealing with missing data. Unfortunately, this approach can be time consuming. This problem is exacerbated when there are many individuals in the pedigree who are not typed at all, as is often the case with large and deep pedigrees. In such a situation it can be computationally impractical to apply the recurrence relation all the way to the founders.
My method uses a different strategy to these problems, and, in the process, can estimate IBD between pairs of individuals with unprecedented speed. Although I start with the same basic recurrence relation above, I expand the probabilistic framework so that it is exactly applicable even when individuals have missing genotype data. The actual implementation allows for greater speed by applying approximations, but now the results are completely deterministic and do not have any Monte Carlo variability. Also, I rework the boundary conditions for the recurrence relation to apply to a specified set of individuals, the “quasi founders.” The quasi founders are, in general, the oldest set of genotyped individuals, although this is not strictly necessary.
This method can estimate the IBD sharing between 11,026 pairs (every possible pair of 148 individuals) from a 13 generation, 3,028 individual pedigree with 700 people genotyped with an average speed of 0.042 seconds with a highly informative marker. (The marker is modeled as a “haplotype marker” composed of many tightly linked SNPs, as might be obtained from a 100k or 500k SNP chip, with 20 alleles and a heterozygosity of 0.87.) The average speed when the marker is a SNP is 0.14 seconds per marker.
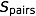
References
Wang T, Fernando R, Vanderbeek S, Grossman M, Vanarendonk J (1995) Covariance between relatives for a marked quantitative trait locus. Genet Sel Evol 27:251–274
Davis S, Schroeder M, Goldin LR, Weeks DE (1996) Nonparametric simulation-based statistics for detecting linkage in general pedigrees. Am J Hum Genet 58:867–880
Updated July 3, 2007